Researchers at Sanford Burnham Prebys have made significant strides in understanding how the three-dimensional structure of chromatin influences gene activity. Published on June 27, 2025, in the journal Genome Biology, this study highlights the complexities of the genome’s architecture, which is often presented in a linear fashion in educational contexts. This simplification can obscure the importance of chromatin’s 3D organization in regulating gene expression.
Human cells contain approximately six feet of DNA tightly packaged within each nucleus. This DNA is organized around proteins called histones, forming a structure known as chromatin. The coiling and looping of DNA into various shapes create regions that interact more closely with one another while isolating others. Disturbances in this 3D configuration have been linked to various diseases, including certain cancers and developmental disorders. Notably, nearly 12% of genomic regions in breast cancer cells exhibit chromatin structural issues.
New Insights into Gene Regulation
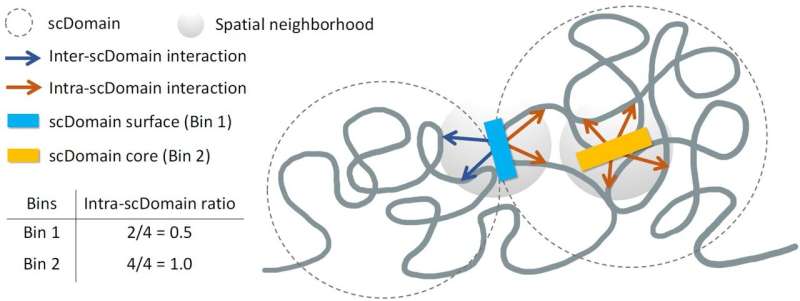
The research team, led by Kelly Yichen Li, Ph.D., a postdoctoral associate at Sanford Burnham Prebys, hypothesized that the 3D shape of genomic regions directly influences gene regulation. “We know that many regions of the genome tend to form what are known as topologically associating domains or TADs,” Dr. Li explained. Within these domains, certain genomic sections interact more frequently, while remaining isolated from others.
During their experiments, the scientists observed that TAD-like regions in individual cells often assumed a globular shape, albeit with irregular contours reminiscent of a variety of potatoes. They proposed that these structural characteristics might affect the function of nearby genes. “If you picture these clumps of chromatin fiber being roughly in the shape of a potato, we predicted that regions of the genome closer to the surface are more active due to exposure to nearby biochemical signals in the cell nucleus,” said Yuk-Lap (Kevin) Yip, Ph.D., a professor and interim director of the Center for Data Sciences at Sanford Burnham Prebys.
Just as a potato’s skin protects its interior, the researchers hypothesized that genes located deeper within a chromatin cluster might be less accessible to signals that promote expression. They developed a method to assess a genomic region’s proximity to the center of a chromatin structure, allowing them to quantify what they termed the “coreness” of these regions. Their findings revealed that surface regions are significantly more active than those located at the core.
Future Research Directions
Dr. Yip noted the potential for this research to advance our understanding of gene activity and its relationship to various diseases across different cell types. “The type of data we can apply this measure to is becoming quite plentiful,” he stated. This study opens avenues for future exploration into how chromatin’s 3D structure may play a role in conditions such as muscular dystrophy.
To further investigate these implications, Dr. Li and Dr. Yip plan to collaborate with the laboratory of Pier Lorenzo Puri, MD, focusing on how the 3D structure of the genome influences muscle stem cell development. Their work promises to enhance our understanding of complex genetic interactions and their role in health and disease.
For more information, refer to the article by Kelly Yichen Li et al titled “Regulatory roles of three-dimensional structures of chromatin domains,” available in Genome Biology (2025). DOI: 10.1186/s13059-025-03659-7.