Researchers from the University of Tokyo have made a groundbreaking discovery in oral microbiome research, revealing the presence of giant DNA elements known as Inocles within human mouths. These previously undetected structures play a vital role in how bacteria adapt to the dynamic environment of the oral cavity. The findings have significant implications for understanding oral health, disease, and the broader field of microbiome research.
This discovery comes amidst a renewed focus on the human microbiome, which includes not only the well-studied gut microbiome but also the oral microbiome. Recent advancements in research techniques have led scientists to uncover previously unknown biological components. Yuya Kiguchi, a Project Research Associate, and his team were motivated by earlier findings of extraneous DNA in soil microbiomes to investigate saliva samples collected by the Yutaka Suzuki Lab at the university.
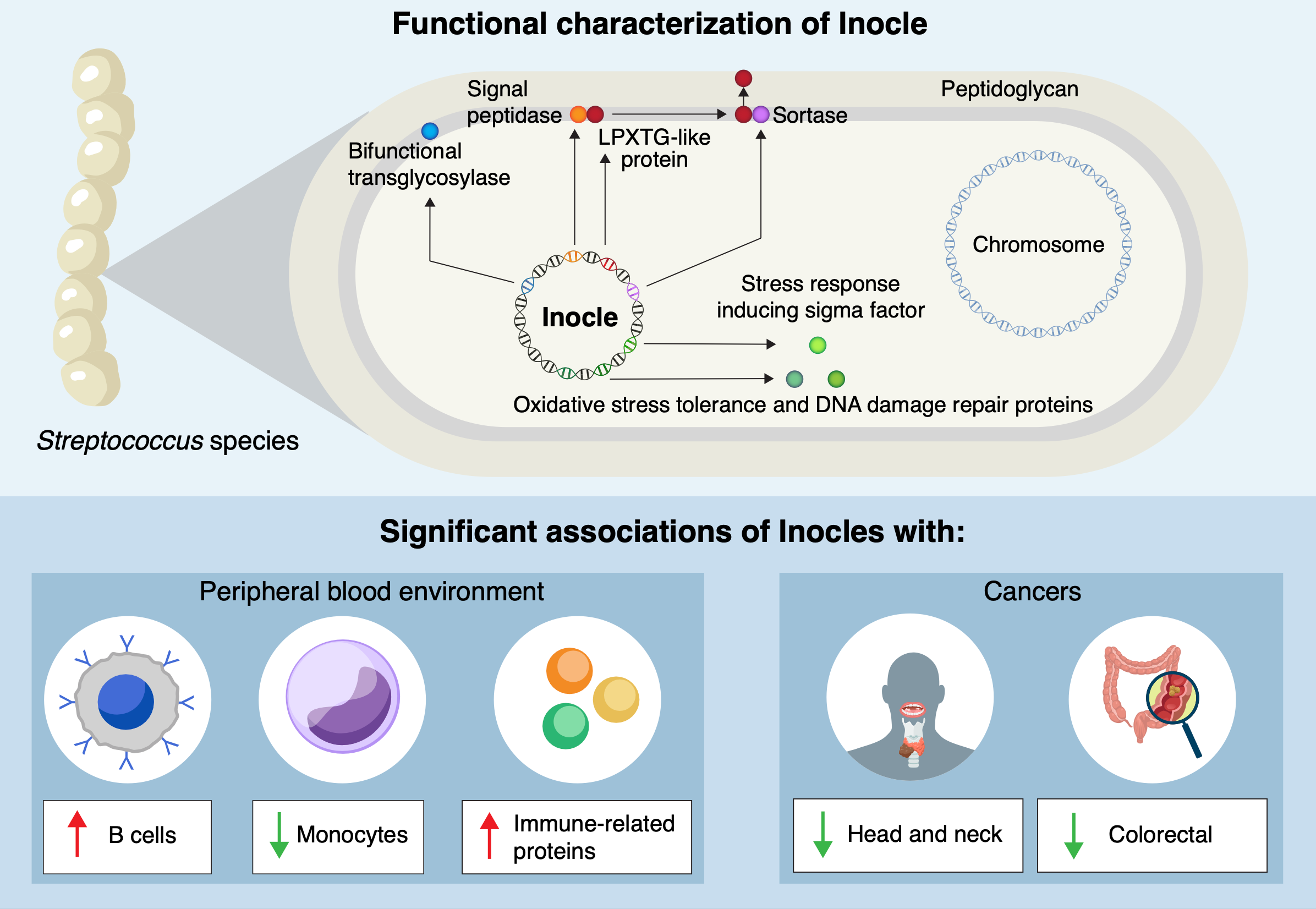
Kiguchi explained, “We know there are a lot of different kinds of bacteria in the oral microbiome, but many of their functions and means of carrying out those functions are still unknown.” Their exploration led to the identification of Inocles, which are examples of extrachromosomal DNA. These DNA fragments exist outside the main genetic material of bacterial cells, akin to finding additional footnotes in a book.
Detecting Inocles posed a challenge due to conventional sequencing methods that fragment genetic data, hindering the reconstruction of large DNA elements. To address this, the research team employed advanced long-read sequencing techniques that can capture extended stretches of DNA. A significant breakthrough was achieved by co-first author Nagisa Hamamoto, who developed a method called preNuc to selectively remove human DNA from saliva samples. This innovation greatly enhanced the quality of sequencing, enabling the assembly of complete Inocle genomes for the first time.
The research revealed that Inocles are primarily associated with the bacteria Streptococcus salivarius. Kiguchi noted, “The average genome size of Inocle is 350 kilobase pairs, making it one of the largest extrachromosomal genetic elements in the human microbiome.” In contrast, other forms of extrachromosomal DNA, known as plasmids, typically measure only a few tens of kilobase pairs.
The size of Inocles equips them with genes that serve various functions, such as resistance to oxidative stress, DNA damage repair, and adaptations to extracellular environments. The research team aims to develop stable methods for culturing bacteria that contain Inocles, which will facilitate further investigations into their functions. This includes whether Inocles can spread between individuals and their potential influence on oral health conditions such as cavities and gum disease.
Despite many Inocle genes remaining uncharacterized, researchers plan to combine laboratory experiments with computational simulations, including tools like AlphaFold, to predict and model the roles these DNA elements may play in oral health. Kiguchi remarked on the significance of their findings, stating, “Given the range of the human population the saliva samples represent, we think 74% of all human beings may possess Inocles.”
This discovery underscores the complexity of the human microbiome, particularly in the oral cavity, where Inocles had previously gone unnoticed due to technological limitations. As research progresses, the potential for Inocles to serve as markers for serious diseases, including cancer, offers a promising avenue for future studies. The ongoing exploration of these giant DNA elements may reshape our understanding of human health and the intricate relationships between humans and their resident microbes.